RNAMoIP
Parameters
Sequence:
A valid RNA sequence as a case-unsensitive
IUPAC-like format. This will be the base of the prediction.
A valid RNA structure that will be take as constraints for the base pairings probabilities prediction.
Pairing levels must be correctly identified, with the following syntax (show ordered by the pairing level): .()[]{}<>
Alpha:
Sets how the RNAMoIP optimize its objectives
Max level:
The maximum depth level of base pairs crossing allowed.
Max Solution Count:
The maximum number of best solutions return by the IP solver.
Information on the alpha parameter
RNAMoIP exposed an alpha parameter that have some agency on what the objective function should maximise.
Lets take a prediction for PDB
5BTP-B as an example.
The parameters will look like the following:
| Alpha |
Predicted Secondary Structure |
VARNA Representation |
| Real Structure |
........((((....[[[[[...((((......)))).....))))............(((..]]]]]..))). |
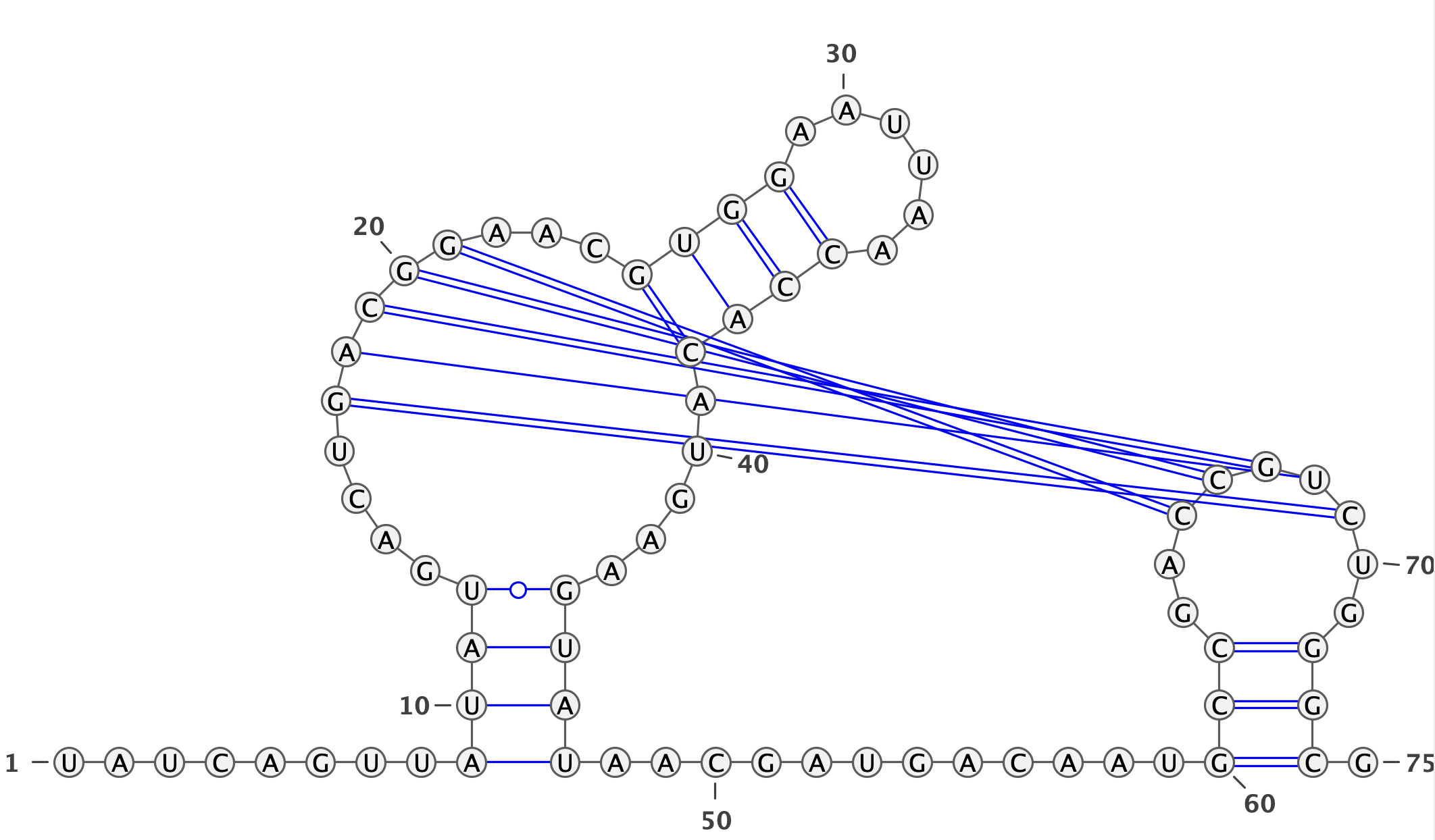 |
| Alpha = 0 |
[[(((((([[[[))))))(((...((((......)))).....]]]].(((.]]....[[[[...))))))]]]] |
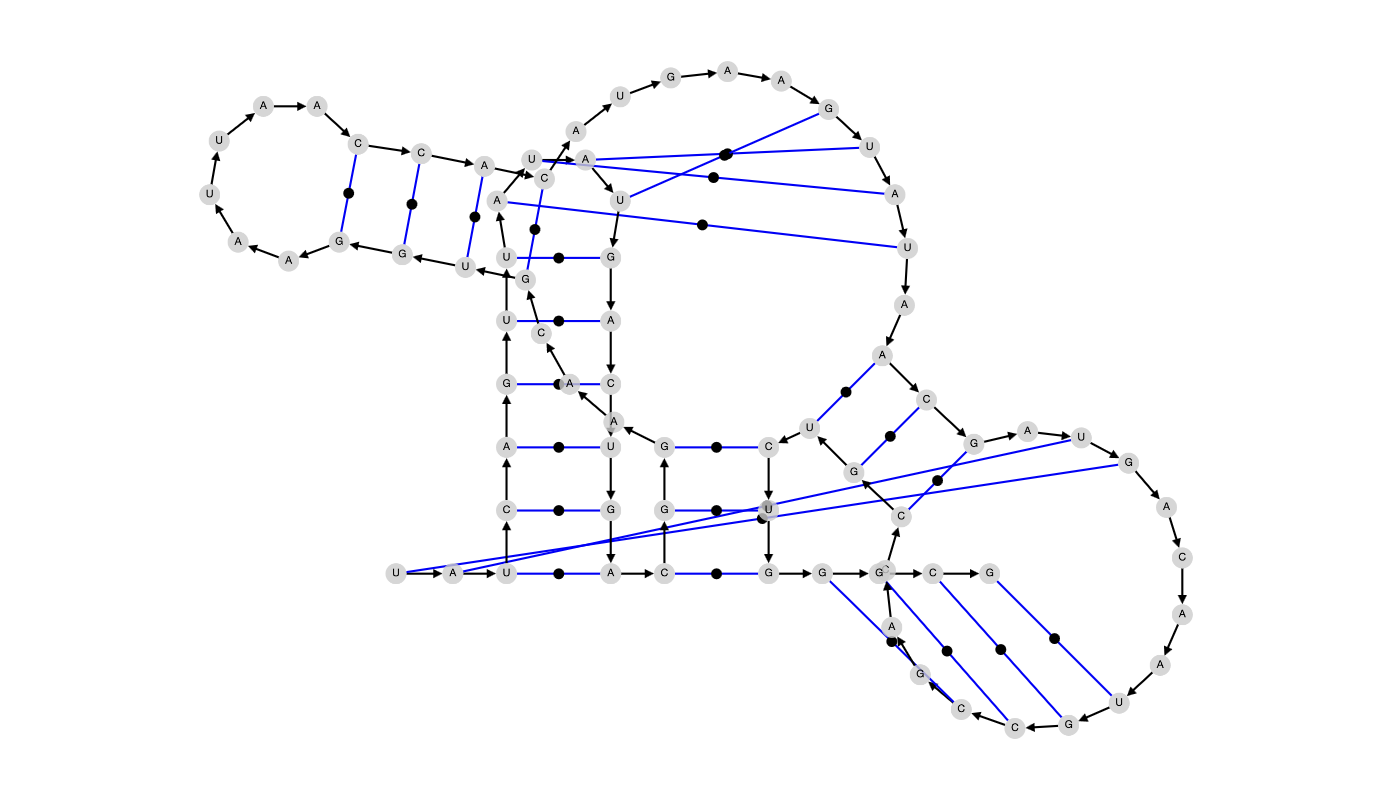 |
| Alpha = 0.1 |
.....((((.((....[[[[....((((......)))).....)).))))........(((....]]]]...))) |
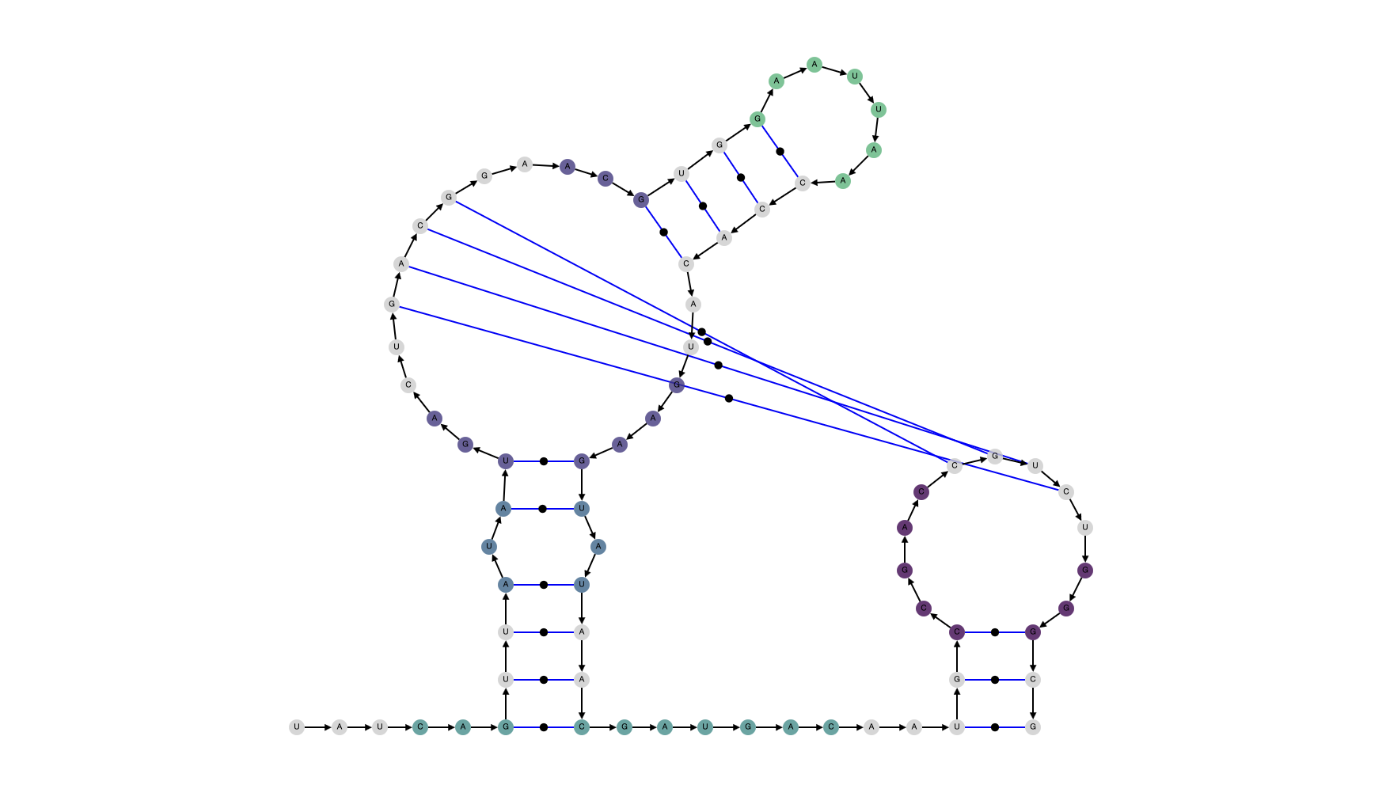 |
| Alpha = 1 |
.....((..........((......(((......)))......))...))........(((...........))) |
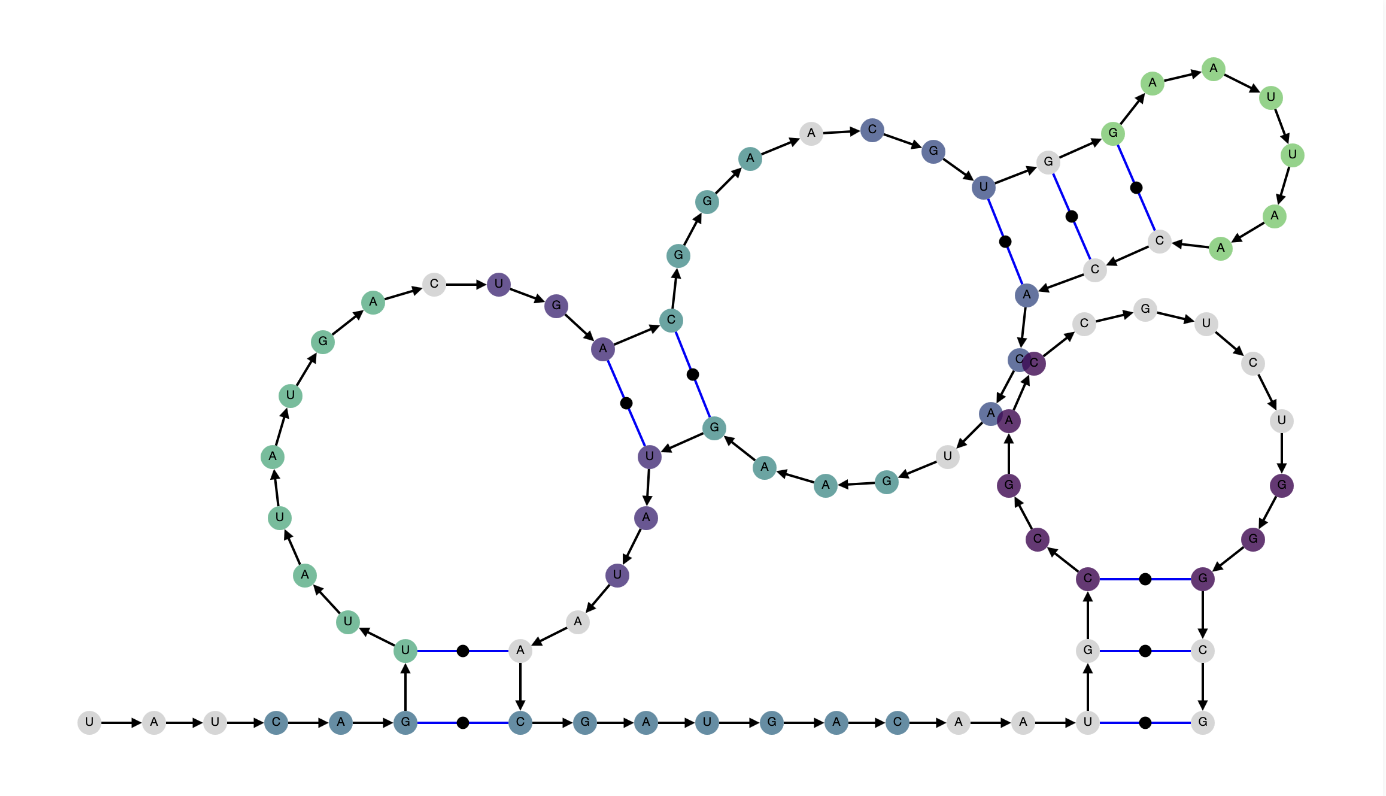 |